Bringing Generative AI Into the EHR: Why DHTI Matter (Part I)
Large Language Models (LLMs) are transforming how we think about clinical decision support, documentation, and patient engagement. Yet despite their impressive capabilities, LLMs have a fundamental limitation that becomes especially important in healthcare: LLMs are stateless. They do not remember prior interactions unless that information is explicitly included in the prompt. For clinical use, this means that patient‑specific data must be added to every prompt if we want the model to generate relevant, safe, and context‑aware output.
This is where the real challenge begins.

Image credit: Grzegorz W. Tężycki, CC BY-SA 4.0 https://creativecommons.org/licenses/by-sa/4.0, via Wikimedia Commons
Why Patient Context Matters for Generative AI in Healthcare
Healthcare workflows depend on rich, longitudinal patient data—medications, allergies, labs, imaging, diagnoses, and more. To generate clinically meaningful output, an LLM must be given this context. Without it, the model is essentially guessing.
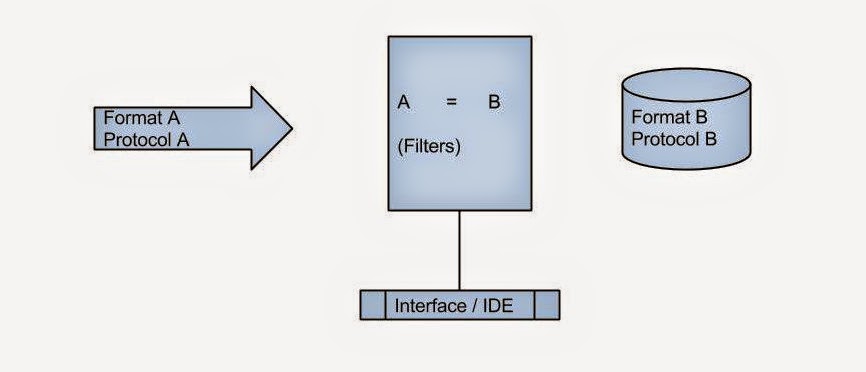
But adding patient data to prompts is not as simple as it sounds. Extracting structured, reliable data from Electronic Medical Records (EMRs) is notoriously difficult. EMRs were not originally designed with AI integration in mind. Data may be siloed, inconsistently structured, or locked behind proprietary interfaces. Even when APIs exist, authentication, authorization, and data‑mapping complexities can slow down innovation.
FHIR: The Standard That Makes Interoperability Possible
Fortunately, the healthcare ecosystem has rallied around a modern interoperability standard: HL7® FHIR® (Fast Healthcare Interoperability Resources). FHIR provides a consistent, web‑friendly way to represent clinical data, making it easier for external applications—including AI systems—to retrieve patient information.
Most major EMRs now expose FHIR APIs that allow authorized systems to query patient‑specific data such as demographics, medications, conditions, and lab results. This shift has been transformative. Instead of custom integrations for each EMR vendor, developers can rely on a shared standard.
FHIR also underpins many modern interoperability frameworks, including SMART on FHIR and CDS‑Hooks. These standards are now widely adopted across the industry, with CDS‑Hooks explicitly designed to connect EMRs to external decision‑support services using FHIR data.
Displaying AI Output Inside the EMR: The Role of CDS‑Hooks
Retrieving data is only half the problem. Once an AI model generates insights, the output must be displayed inside the clinician’s workflow—not in a separate window, not in a separate app, and not in a place where it will be ignored.
This is where CDS‑Hooks comes in.
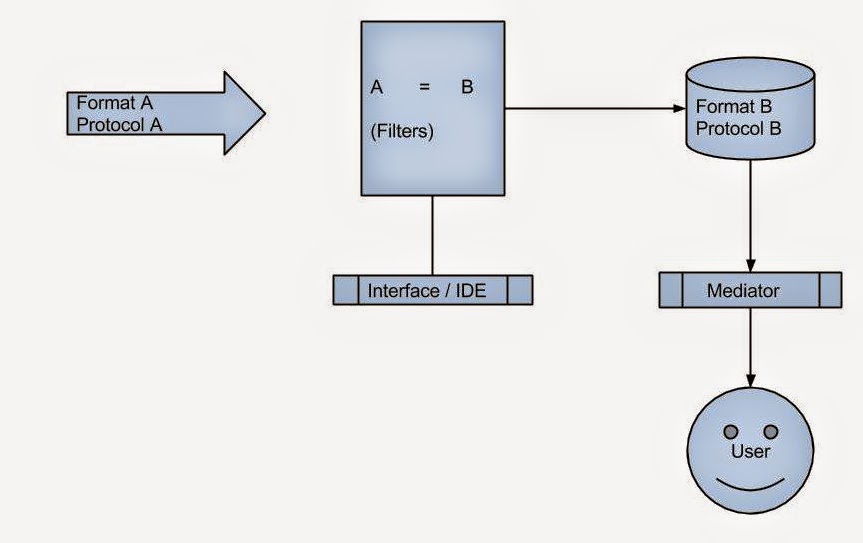
CDS‑Hooks is a standard that allows EMRs to call external decision‑support services at specific points in the clinical workflow. When a clinician opens a chart, writes an order, or reviews a medication list, the EMR can trigger a “hook” that sends key context—including the patient ID—to a backend service. That backend can then use FHIR APIs to retrieve the necessary patient data, run AI models, and return actionable “cards” that appear directly inside the EMR interface.
This pattern is powerful because:
- It keeps clinicians in their workflow
- It ensures AI output is tied to real‑time patient context
- It avoids sending large amounts of PHI directly from the EMR to the AI model
In short, CDS‑Hooks is the bridge between EMRs and modern AI‑powered decision support.
DHTI: A Reference Architecture for GenAI in Healthcare
As interest in generative AI grows, developers and researchers need a framework that brings all these pieces together—LLMs, FHIR, CDS‑Hooks, EMR integration, and modular AI components. DHTI (Distributed Health Technology Interface) is one such open‑source project.
DHTI embraces the standards that matter:
- FHIR for structured data exchange
- CDS‑Hooks for embedding AI output in the EMR
- LangServe for hosting modular GenAI applications
- Ollama for local LLM hosting
- OpenMRS as an open‑source EMR environment
The project’s documentation highlights how CDS‑Hooks is used to send patient context (including patient ID) and how backend services retrieve additional data using FHIR before generating AI‑driven insights. DHTI’s architecture is intentionally modular, allowing developers to prototype new GenAI “elixirs” (backend services) and UI “conches” (frontend components) that plug directly into an EMR environment.
You can explore the project here:
Why This Matters for the Future of Clinical AI
Healthcare AI must be:
- Context‑aware
- Integrated into clinical workflows
- Standards‑based
- Secure and privacy‑preserving
- Interoperable across EMRs
LLMs alone cannot meet these requirements. But LLMs combined with FHIR, CDS‑Hooks, and frameworks like DHTI can.
This is how we move from isolated AI demos to real, production‑ready clinical tools. Try DHTI and Help Democratize GenAI in Healthcare