The clinical context
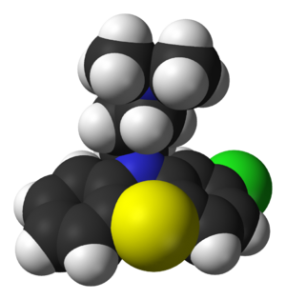
| Space-filling model of the chlorpromazine molecule. X-ray diffraction data from H. S. Yathirajan, M. A. Ashok, B. Narayana Achar and M. Bolte (April 2007). “Chlorpromazinium picrate”. Acta. Cryst. E63 (4) : o1795-o1797. DOI:10.1107/S1600536807011555. (Photo credit: Wikipedia) |
I recently read an article titled “Structural, phylogenetic and docking studies of D-amino acid oxidase activator (DAOA), a candidate schizophrenia gene”. (Open access, link to full text below).
This is a typical example of how over-zealous bioinformatics analysis without a clinical context can churn out garbage. The article after multiple analysis concludes that DAOA interacts with DAO. Well that is why DAOA is named DAO activator. The article also predicts that a predicted ligand of the predicted tertiary structure could be a schizophrenia drug.
If only it were all so simple! If Only….
Ref: http://www.tbiomed.com/content/10/1/3