Clinical knowledge representation for reuse
The need for computerized clinical decision support is becoming increasingly obvious with the COVID-19 pandemic. The initial emphasis has been on ‘replacing’ the clinician which for a variety of reasons is impossible or impractical. Pragmatically, clinical decision support systems could provide clinical knowledge support for clinicians to make time-sensitive decisions with whatever information they have at the point of patient care.
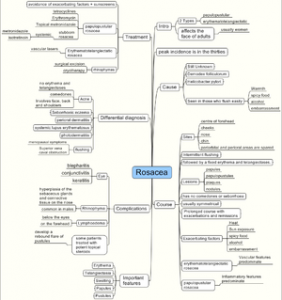
Providing clinical decision support requires some formal way of representing clinical knowledge and complex algorithms for sophisticated inference. In knowledge management terms, the information requires to be transformed into actionable knowledge. Knowledge needs to be represented and stored in a way conducive to easy inference (knowledge reuse)1. I have been exploring this domain for a considerable period of time, from ontologies to RDF datasets. With the advent of popular graph databases (especially Neo4J ), this seems to be a good knowledge representation method for clinical purposes.
To cut a long story short, I have been working on building a suite of JAVA libraries to support knowledge extraction, annotation and transformation to a graph schema for inference. I have not open-source it yet as I have not decided on what license to use. However, I am posting some preliminary information here to assess interest. Please give me a shout, if you share an interest or see some potential applications for this. As always, I am open to collaboration.
The JAVA package consists of three modules. The ‘library’ module wraps the NCBI’s E-Utils API to harvest published article abstracts if that is your knowledge source. Though data extraction from the clinical notes in EMR’s is a recent trend, it is challenging because of unstructured data and lack of interoperability. The ‘qtakes’ module provides a programmable interface to my quick-ctakes or the quarkus based apache ctakes, a fast clinical text annotation engine. Finally, the graph module provides the Neo4J models, repositories and services for abstracting as a knowledge graph.
The clinical knowledge graph (ckb) consists of entities such as Disease, Treatment and Anatomy and appropriate relationships and cypher queries are defined. The module exposes services that can be consumed by JAVA applications. It will be available as a maven artifact once I complete it.
UPDATE: May 30, 2021: The library (ckblib) is now available under MPL 2.0 license (see below). Feel free to use it in your research.
- 1.Toward a Theory of Knowledge Reuse: Types of Knowledge Reuse Situations and Factors in Reuse Success. Journal of Management Information Systems. Published online May 31, 2001:57-93. doi:10.1080/07421222.2001.11045671